10 6 Using Latent Block Models to Detect Structure in Ecological Networks 6.1 Introduction 6.2. Formalism 6.3. Probabilistic mixture models for networks 6.4. Statistical inference 6.5. Application 6.6. Conclusion 6.7. References
11 7 Latent Factor Models: A Tool for Dimension Reduction in Joint Species Distribution Models 7.1. Introduction 7.2. Joint species distribution models 7.3. Dimension reduction with latent factors 7.4. Inference 7.5. Ecological interpretation of latent factors 7.6. On the interpretation of JSDMs 7.7. Case study 7.8. Conclusion 7.9. References
12 8 The Poisson Log-Normal Model: A Generic Framework for Analyzing Joint Abundance Distributions 8.1. Introduction 8.2. The Poisson log-normal model 8.3. Data analysis: marine species 8.4. Discussion 8.5. Acknowledgments 8.6. References
13 9 Supervised Component-Based Generalized Linear Regression: Method and Extensions 9.1. Introduction 9.2. Models and methods 9.3. Case study: predicting the abundance of 15 common tree species in the forests of Central Africa 9.4. Discussion 9.5. References
14 10 Structural Equation Models for the Study of Ecosystems and Socio-Ecosystems 10.1. Introduction 10.2. Structural equation model 10.3. Case study: biodiversity in managed forests 10.4. Discussion 10.5. Acknowledgments 10.6. References
15 List of Authors
16 Index
17 End User License Agreement
1 Introduction Table I.1. Chapters and contents
2 Chapter 1 Table 1.1. Evolution of model selection criteria (AIC and ICL) as a function of ...
3 Chapter 3Table 3.1. Prevalence of hybrids: observed and estimated using the Viterbi algor...
4 Chapter 4Table 4.1. Parameters estimated by maximum likelihood
5 Chapter 5Table 5.1. Notation of variables in the MHMM-DFTable 5.2. Interpretation of parameters of the MHMM-DF based on binomial distrib...Table 5.3. Boundaries of the five abundance classes for the seven weed species i...Table 5.4. Probabilities of survival, colonization and germination from a dorman...Table 5.5. BIC selection values for models with and without taking account of cr...Table 5.6. Probabilities of survival and emergence from a dormant state in the M...
6 Chapter 8Table 8.1. Glossary of species codes, scientific names and common names for spec...
7 Chapter 9Table 9.1. Spearman correlations between observed and predicted abundances obtai...
8 Chapter 10Table 10.1. Variables used in the case studyTable 10.2. Latent variables based on prior knowledge
1 Chapter 1 Figure 1.1. The map at the top shows the tracking data for a male Cape dolphin (... Figure 1.2. Figure extracted from Figure 4 in Lopez et al. (2015). The black lin... Figure 1.3. Graphical model. For a color version of this figure, see www.iste.co... Figure 1.4. Illustration of the quantities present in equations [1.5]–[1.8]. Pt ... Figure 1.5. Masked Booby (Sula dactylatra) Photo: Sophie Bertrand. For a color v... Figure 1.6. Area of study (shown in red on the map) and three trajectories obtai... Figure 1.7. Result of Kalman smoothing on part of the booby trajectories. Smooth... Figure 1.8. Representation of states along trajectories estimated using two diff... Figure 1.9. Distribution of our chosen metrics for the states estimated using ou... Figure 1.10. Contingencies of estimated states for our two models. For a color v... Figure 1.11. Evolution of the probability of being in state 1 or state 2 over ti... Figure 1.12. Evolution of estimated transition probabilities as a function of di... Figure 1.13. Study zone (red dot on the map) and three trajectories of three dif...
2 Chapter 2 Figure 2.1. Illustration of van Noordwijk and de Jong’s (1986) “Y” model. Exampl... Figure 2.2. Directed acyclic graph of the model. The squares represent observabl... Figure 2.3. A posteriori distributions of parameters in the latent model (logari...Figure 2.4. Comparison of observed and simulated ring width from 1989 to 2015 an...Figure 2.5. Boxplot of resource (net primary productivity, in gC.m−2.year−1) sim...Figure 2.6. Illustration of the developed Bayesian model, with process and data ...Figure 2.7. Correlation between sinks and probabilities. a) Correlation density ...
3 Chapter 3Figure 3.1. Schematic illustration of a hidden Markov modelFigure 3.2. Two-state capture–recapture model expressed in HMM formFigure 3.3. Multi-state capture–recapture model expressed in HMM formFigure 3.4. Diagram of a dynamic occupancy model expressed as an HMMFigure 3.5. Identification of local minima in the 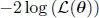 deviance of an HMM. Numerica...Figure 3.6. Visualization of heterogeneity: map of the heterogeneity class to wh...
deviance of an HMM. Numerica...Figure 3.6. Visualization of heterogeneity: map of the heterogeneity class to wh...
4 Chapter 4Figure 4.1. Expectation of the total number of cases associated with the posteri...Figure 4.2. Joint posterior distributions of couples (α, κ), (t0, α) and (t0, κ)...Figure 4.3. Map of wolf detections in southeastern France (black dots) and the a...Figure 4.4. Estimated response curves. Estimated relations between individual de...Figure 4.5. Predicted occupation probability map for 2016, obtained using the mo...Figure 4.6. Proportion of plants susceptible to WMV across the area of study. Fo...Figure 4.7. Proportions of classic and invasive variants in a landscape: data an...
5 Chapter 5Figure 5.1. Illustration of dependency relationships in an MHMM-DF. Case of two ...Figure 5.2. Layout of the 10 fields and 90 patches in the experimental farm at E...
6 Chapter 6Figure 6.1. The Chilean network: 1,362 trophic interactions observed in the inte...Figure 6.2. Adjacency matrix and corresponding representation of the non-directe...Figure 6.3. Incidence matrix and corresponding representation of the bipartite b...Figure 6.4. Simulation of a modular network for the parameters shown on the left...Figure 6.5. Simulation of a food web for the parameters shown on the left. A rea...Figure 6.6. Simulation of a nested bipartite network for the parameters shown on...Figure 6.7. Simulation of a bipartite network with a modular and nested structur...Figure 6.8. Schematic representation (based on Picard et al. 2009) of the estima...Figure 6.9. Classic representation obtained using the R bipartite package. Diffe...
7 Chapter 7Figure 7.1. The three factors that determine the actual distribution of a specie...Figure 7.2. Localization and names of the 18 gradients of ORCHAMP. For a color v...Figure 7.3. Effective sample size (ESS, top panels) and potential scale reductio...Figure 7.4. Distribution of TSS and RMSE score across species for in-sample pred...Figure 7.5. Posterior support values for species regression coefficients. Red if...Figure 7.6. The residual correlation matrix. Only significant values (i.e. 95% c...Figure 7.7. Model-based ordination analysis. The two latent variables can be see...Figure 7.8. Model-based ordination analysis, as above, but when we include the h...Figure 7.9. Cross-validation predicted probability of (a) presence and (b) cross...
8 Chapter 8Figure 8.1. Illustration of dependency in the PLN model. Random variables are ci...Figure 8.2. PLN: geometric view of the model for two species. A) Positions in th...Figure 8.3. Poisson log-normal model with the “site” covariate. Representation o...Figure 8.4. Poisson log-normal model with the “period” covariate. Representation...Figure 8.5. Dimension reduction for (left to right): a model without covariates;...Figure 8.6. Representation of samples on the AEI islet in the first principal pl...Figure 8.7. Effect of parameter λ on edge density (left) and model fitting (righ...Figure 8.8. Stability of edges selected for the model including the effects of t...Figure 8.9. The selected interaction network: visualization using the PLNmodels ...Figure 8.10. The selected interaction network: visualization of the partial corr...Figure 8.11. Interactions between two sea urchin species: red (STRFRAAD, Strongy...
Читать дальше

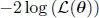 deviance of an HMM. Numerica...Figure 3.6. Visualization of heterogeneity: map of the heterogeneity class to wh...
deviance of an HMM. Numerica...Figure 3.6. Visualization of heterogeneity: map of the heterogeneity class to wh...










