Genotyping by Sequencing for Crop Improvement
Здесь есть возможность читать онлайн «Genotyping by Sequencing for Crop Improvement» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Genotyping by Sequencing for Crop Improvement
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:4 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 80
- 1
- 2
- 3
- 4
- 5
Genotyping by Sequencing for Crop Improvement: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Genotyping by Sequencing for Crop Improvement»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
A thoroughly up-to-date exploration of genotyping-by-sequencing technologies and related methods in plant science Genotyping by Sequencing for Crop Improvement,
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Genotyping by Sequencing for Crop Improvement», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
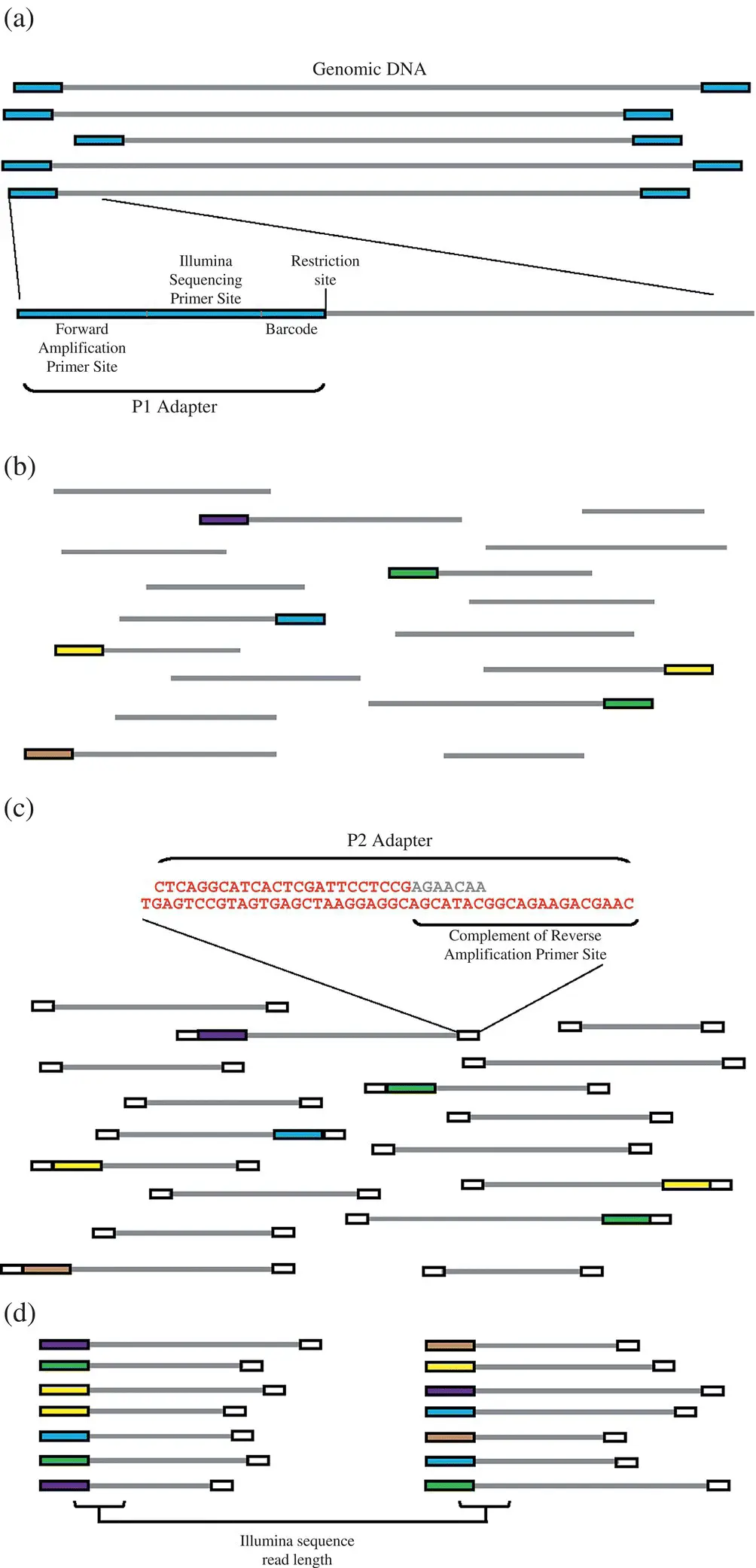
Figure 2.3 Illustration of various steps involved in the generation of RAD‐based SNP marker generation. (a) Ligate P1 adopter to digested genomic DNA. (b) Poor barcoded sample and shear. (c) Ligate P2 adopter to sheared fragments. (d) selective amplification of RAD tags.
Source: The figure is reproduced from Baird et al. (2008) available with Creative Commons Attribution License (CC BY).
An IFC is primed in the IFC Controller before loading of samples and assays. The IFC controller pressurizes the control lines and closes the interface valves. After priming of IFC, samples and Taqman assays are loaded into the IFC and then again put into the controller for the mixing of sample and assays into the reaction chambers. In the loading process, on application of pressure, fluid from the sample inlets and assay inlets are pushed into the corresponding fluid lines. The chip utilizes carry over slug design (CS) which allows precise metering and mixing of fluids in a reaction (Unger et al. 2000) chamber (nanolitres of samples and assays are used in a reaction). The loading process takes 45 minutes in the controller and after that, the IFC is being loaded into the standalone PCR and the reaction starts in reaction chambers by thermal cycling in presence of fluorophore‐containing probes. After completion of PCR, the data are acquired in the EP1 fluorescence system and the data analysis is done using bioinformatic tools and Software.
The processing of plant samples, i.e. DNA requires preamplification, which is done using specific‐target amplification and locus‐specific primers. After diluting the preamplified products with distilled water, PCR amplification with allele‐specific primers is carried out (Kishora et al. 2020). During PCR amplification, two fluorogenic probes corresponding to one of the two alleles in a biallelic SNP are used to probe the SNP site. The probes are made up of a fluorescent reporter dye attached to the probe's 5′ end and a nonfluorescent quencher attached to the probe's 3′ end. The reporter dye's fluorescence is muted when the probe is intact because the reporter dye is close to the quencher. The probe anneals selectively to a complementary sequence during PCR. This causes the probe to cleave, releasing the reporter dye, resulting in increased fluorescence with each PCR cycle. This increase in fluorescence occurs only if the target sequence is complementary to the probe and is amplified during PCR (Seeb et al. 2009).
MALDI TOF‐based SNP Genotyping‐Sequenom
MALDI MS (matrix‐assisted laser desorption/ionization mass spectrometry) allows for the quick study of high molecular weight biomolecules (e.g. DNA, proteins, peptides, polysaccharides, and other organic compounds) and other chemical substances (e.g. polymers, macromolecules, and hybrid materials). It produces single‐charged ions and has a high detection accuracy and sensitivity. Sequenom incorporates matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectrometry technique for highly accurate, low‐throughput SNP genotyping. MALDI‐TOF‐based SNP genotyping platforms take the advantage of differences in mass between allele‐specific primer and extension products. During the MALDI process, analyte molecules are mixed with low molecular weight organic molecules like α‐cyano‐4‐hydroxycinnamic acid (CHCA) and 2,5‐dihydroxybenzoic acid (DHB) called matrix and the mixture is crystallized when dried over SpectroCHIP. Then the crystallized matrix‐analyte mixture (a large quantity of matrix is used) is irradiated with a laser beam with an emitting wavelength range in UV or mid IR range. The irradiation causes desorption and ionization of analyte molecules in the form of singly protonated ions. The system uses a TOF mass analyzer. The protonated ions are then accelerated with uniform translational energy on application of the electric field. Then the charged ions are moved to the drift region which is free of electric field. The velocity of charged ions in the drift region is determined by the mass to charge ratio of the ions which in turn is calculated by measuring time required by the ions to travel through the flight tube ( Figure 2.4).
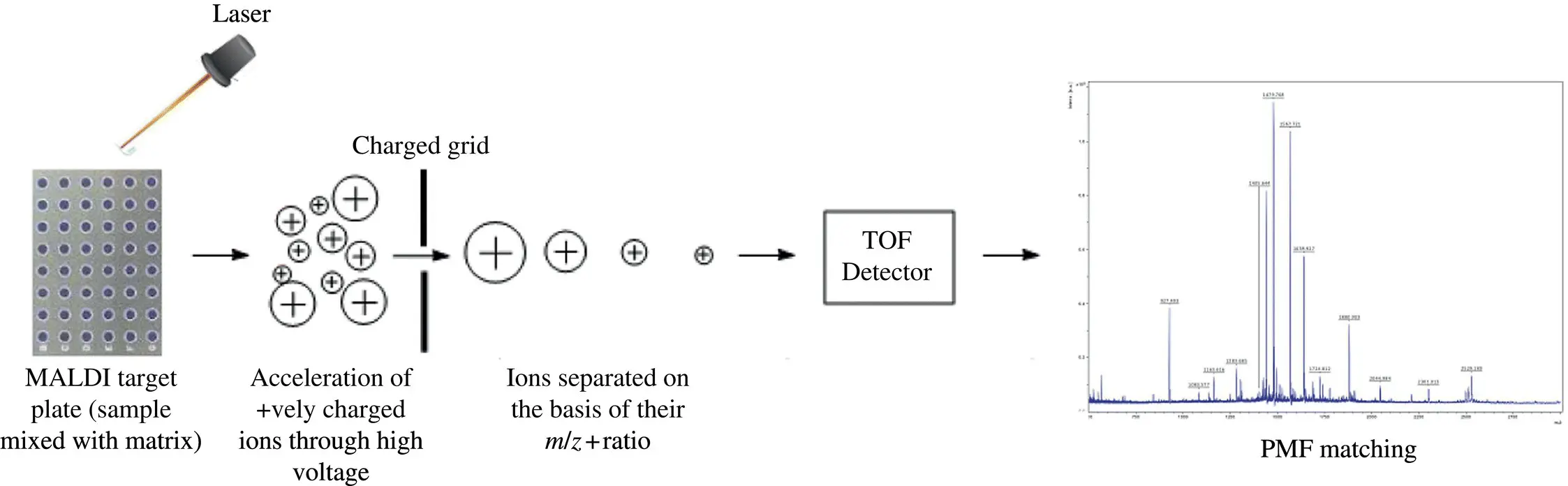
Figure 2.4 Schematic illustration of work‐flow in a MALDI‐TOF MS
Source: The figure is reproduced from Singhal et al., 2015, figure 01(p.03), available with Creative Commons 4.0
For SNP genotyping, the target regions of the sample (5–10 ng) are amplified for each assay within the multiplex. The PCR product is mixed with the shrimp alkaline phosphatase (SAP) which dephosphorylates any residual dNTPs, which might interfere with the allele‐specific termination at the later stage. The amplification process is followed by a post‐PCR primer extension where an allele‐specific primer is annealed at the adjacent position of the SNP site. The extension reaction cocktail contains four terminating nucleotides (ddNTPs). The extension involves the addition of a single nucleotide and hence alleles are differentiated by the molecular mass of the extension product. The extension products are then conditioned with the ion‐exchange resin and a nano dispensing device loads a few nanoliters of sample product to the prefabricated matrix loaded chip array (384‐pad spectroCHIP) (Oath et al. 2009). The SpectroCHIP is then loaded to the MALDI‐TOF MS analyzer for the analysis.
2.2.3 Custom Assay Technologies
Once any significant SNP with a vital role in gene regulation, phenotype change, disease response, and so on is discovered, the next step is to validate it in a larger population. Arrays designed with few vital SNPs, mainly for validation purposes are called custom‐designed SNP assays. These assays must meet at least majority of the following criteria:
1 should be able to analyze large amounts of samples quickly,
2 should have a high design to assay conversion rate for custom SNPs, and
3 should be reliable and cost‐effective.
To achieve these requirements, these technologies have incorporated a variety of sophisticated molecular biology techniques. The advantage of customized array is that SNPs from genomic regions of interest can be specifically added to the array and the number of interested SNPs to be fabricated on the array can be adjusted to the customer's needs. Customized SNP array cost per sample is low to moderate approximately $28–$90 (USD) while the NGS method have approximately $35 per sample price for GBS (Peng et al. 2017) which will increase drastically from diploid to polyploid as much higher coverage is required for accurate SNP calling. The major disadvantage of customized SNP array is that it requires prior genomic information and location (Vos et al. 2015). Furthermore, its design and further optimization can take a long time. Another issue is that SNP discovery requires fewer samples, which will primarily remove rare alleles while capturing common alleles (Gravel et al. 2011).
2.2.4 Summary
SNPs constitute one of the most popular and significant genetic markers in studying disease development and progression, breeding crops for improved traits etc. Two major strategies are involved in SNP genotyping viz. allele discrimination and allele detection. The high‐throughput platforms used for SNP genotyping have generated huge amount of data in many crops. Further utilization of this information in plant breeding is important for crop improvement.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Genotyping by Sequencing for Crop Improvement»
Представляем Вашему вниманию похожие книги на «Genotyping by Sequencing for Crop Improvement» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Genotyping by Sequencing for Crop Improvement» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












