Genotyping by Sequencing for Crop Improvement
Здесь есть возможность читать онлайн «Genotyping by Sequencing for Crop Improvement» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Genotyping by Sequencing for Crop Improvement
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:4 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 80
- 1
- 2
- 3
- 4
- 5
Genotyping by Sequencing for Crop Improvement: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Genotyping by Sequencing for Crop Improvement»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
A thoroughly up-to-date exploration of genotyping-by-sequencing technologies and related methods in plant science Genotyping by Sequencing for Crop Improvement,
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Genotyping by Sequencing for Crop Improvement», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
In the last couple of years, the detection and exploitation of DNA polymorphisms with molecular markers and their utilization in breeding programs have transformed the pace and accuracy of these experiments. Ultimately, a complete shift from phenotype‐based breeding to molecular breeding has been evident. With the advancement of NGS technology, there is a rapid rise in the application of SNP‐based genotyping. The popularity of these programs depends on the cost incurred as well as the ease of their application. Also, these SNP‐based genotyping platforms offer many vital advantages over other marker systems like reproducibility, high marker density, automation, and many more. Many modern breeding techniques, such as genome‐wide association studies (GWAS) and genomic selection (GS), necessitate a large number of markers, which is difficult to achieve using PCR‐based markers like SSR.
2.2 SNP Genotyping Platforms
Accurate detection of SNP is the first and foremost step for its mainstream application in breeding experiments. Besides the sequencing techniques used, reads length, coverage, and robust assembly approach are the control points for sequencing errors. Sequencing technologies with long reads length (in kb’s) like PacBio single‐molecule real‐time (SMRT) sequencing (Eid et al. 2009), Illumina TruSeq synthetic long‐read technology (McCoy et al. 2014), and Oxford nanopore sequencing (Branton et al. 2010) are available to develop the complete and correct reference genome assembly of any organism, which sort out many of the problems in SNP discovery. Not only in genome sequencing, these long‐read methodologies have been successfully deployed for SNP mining and provide a more precise understanding of the complexity of isoforms for organisms lacking a reference (Piriyapongsa et al. 2018). Once a reference genome is available, the application of SNPs in its breeding program becomes comparatively easier. A concise layout for SNP‐based breeding experiment is provided ( Figure 2.1).
2.2.1 SNP Genotyping Versus SNP Discovery
First, a diverse set of genotypes are selected based on phenotype for the creation of SNP panel with the concept that as these genotypes are hypervariable in several phenotypes, the same should reflect from their sequences. The reference genome sequence is used for reference‐based assembly of the reads generated from sequencing of these genotypes (S1–S10 in Figure 2.1). A set of hypervariable loci are filtered through some statistical analysis. Once discovered, these can be genotyped in any other accession, which is known as SNP genotyping, as genotyping is the process of knowing the allelic status of an organism at a particular locus. The process till the identification of hypervariable loci is known as SNP discovery. The discovery of SNPs in crops prompted the development of the simple and low‐cost genotyping platforms. A flowchart related to different SNP genotyping technologies has been provided ( Figure 2.2).
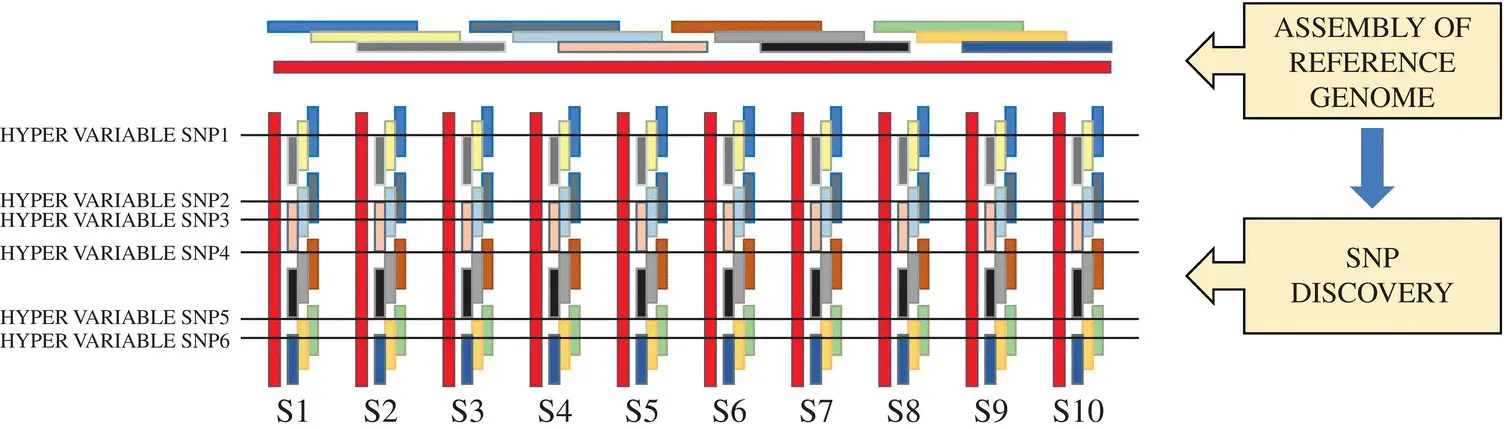
Figure 2.1 A pipeline for SNP discovery (S1–S10 are different diverse accession of the same species to which the reference belongs).
2.2.2 Types of SNP Genotyping Platforms
Although multiple classification systems are based on the reaction, fluorescence used, PCR usage, etc., but broadly all SNP genotyping methods can be categorized into two groups based on the detection mechanism, i.e. Allelic discrimination and Allelic detection. Both of these groups can be further classified based on the reaction chemistry and other variables.
2.2.2.1 Allelic Discrimination
Allelic discrimination depends on allele‐specific biochemical reactions where the alternate alleles are discriminated based on their extension (primer extension methods), hybridization (array for both alleles), and differential enzymatic cleavage pattern. A broader classification for different allelic discrimination method is mentioned below:
2.2.2.1.1 PCR‐Free Genotyping Technology
Among the conventional molecular markers, RFLP is based on the detection of mutation at the restriction site which is usually SNP. Except for RFLP, which relies on restriction digestion followed by detection of digested DNA via southern blotting, other majority of the SNP genotyping methods require amplification of the SNP containing genomic region prior to polymerase chain reaction (PCR). This preamplification step is inevitable from most of the genotyping techniques, despite the fact that the PCR process is relatively expensive.
Invader Assay
Several PCR‐free genotyping methods have been developed to date, one of such methods is invader assay (Lyamichev et al. 1999). Invader assay is based on nucleotide‐specific cleavage by a structure‐specific “flap” endonuclease, in the presence of an invading oligonucleotide. This reaction is followed by a subsequent secondary reaction that generates allele‐specific signals using fluorescence resonance energy transfer (FRET) oligonucleotide cassettes. Both of the reactions in invader assay are a “single vessel reaction” as well as isothermal reaction, hence are easily automatable. Although the whole process is highly accurate and automatable, the need of a large amount of DNA is one of its major drawbacks which can be omitted by coupling the reaction with PCR. Many other PCR‐free methodologies have been proposed like padlock probe ligation (Nilsson et al. 1994) the rolling circle DNA amplification (RCA) process (Baner et al. 1998). Although these gel‐free methods can be automated, and are highly accurate, their application is limited to only a small number of SNPs, hence genotyping of large numbers of SNPs covering the whole genome for genomic assisted breeding is very difficult.
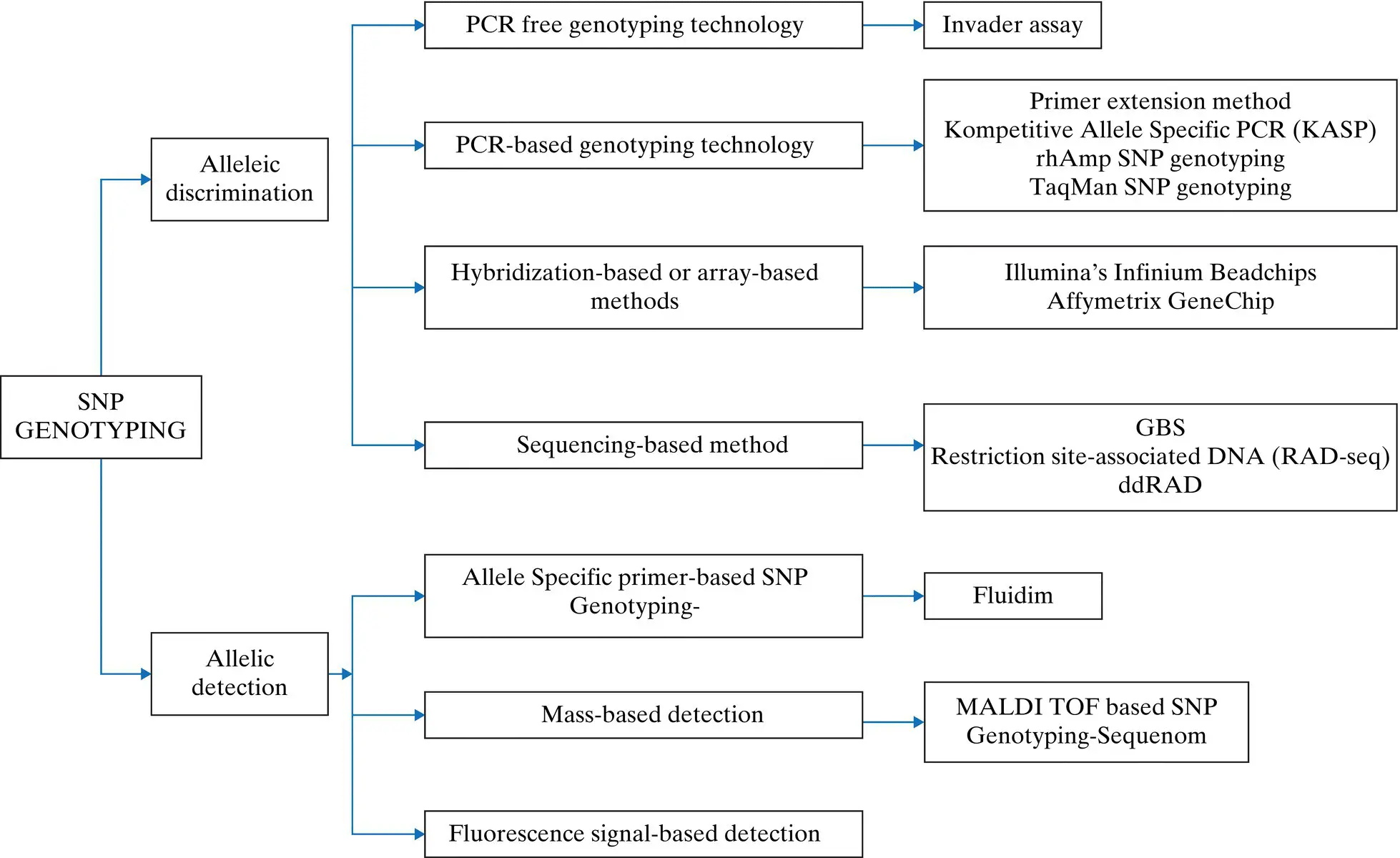
Figure 2.2 A schematic representation of different SNP genotyping technologies.
The fluorescence‐based detection systems were further advanced and were designed in such a manner to detect change in polarization of fluorescence which is caused by the decreased mobility of a fluorophore due to an increase in molecular mass. During SNP genotyping with this method, the allele‐specific incorporation of fluorescently labeled ddNTPs can be detected as an increase in polarized fluorescence (Chen et al. 1999). As this amendment does not require fluorescently labeled primer and also a simple detection platform for genotyping is required, so the cost incurred is decreased. The addition of one extra step (PCR and post‐PCR processing steps) compared to single‐step methods makes it more complicated. Further, these fluorescence polarization‐based methods are usually unsuitable for high‐throughput genotyping because of the lack of multiplexing capability.
2.2.2.1.2 PCR‐based Detection System
Primer Extension Method
PCR‐based primer discrimination method simply exploits the accuracy of polymerase to exclude nonspecific nucleotides during extension. In this method, two forward primers are designed from the locus differing in the genotypes while a common reverse primer is used. Both forward primers are specific for their respective alleles. Two different reactions are set for a sample where each reaction contains a different forward primer. Amplification will take place only in any one of these reactions otherwise both DNA is homozygous for the concerned loci.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Genotyping by Sequencing for Crop Improvement»
Представляем Вашему вниманию похожие книги на «Genotyping by Sequencing for Crop Improvement» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Genotyping by Sequencing for Crop Improvement» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












