DNA Origami
Здесь есть возможность читать онлайн «DNA Origami» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:DNA Origami
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:5 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 100
- 1
- 2
- 3
- 4
- 5
DNA Origami: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «DNA Origami»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
Discover the impact and multidisciplinary applications of this subfield of DNA nanotechnology DNA Origami
DNA Origami
DNA Origami
DNA Origami — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «DNA Origami», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Figure 1.1 History of DNA nanotechnology and DNA origami technology. Progress of DNA nanotechnology and DNA origami technology and major findings and inventions in this field.
1.1.1 DNA Nanotechnology Before the Emergence of DNA Origami
For construction of a large‐sized DNA nanostructure by self‐assembly, rigid DNA building blocks are required. The first DNA building block, the double‐crossover (DX) motif, is one of the most essential and important inventions in DNA nanotechnology [7]. In the DX motif, two double‐stranded DNAs (dsDNA) are connected at two crossover points in parallel and antiparallel arrangements, which reduces the flexibility of the single dsDNA ( Figure 1.2c). The two crossover points are separated by a defined number of base pairs. Using these DX tiles as building blocks, large nanostructures can be constructed via hybridization of the four sticky ends introduced to the DX tiles, which directs the self‐assembly into 2D nanostructures [10]. By using this strategy, 2D building blocks have been further developed for the preparation of various 2D tiles, such as triple‐crossover [11], triangular [12, 13], and 4 × 4 tiles [14]. This concept has also been extended to double‐helix bundled building blocks designed for the construction of tubular structures [15]. All the structures were constructed by simply using defined numbers of unmodified DNA strands. For further extension of the nanostructures, a more complicated design of the building blocks and sequences with larger numbers of DNA strands are needed.
In addition, mechanical DNA nanomachines with a controllable molecular system were developed. An extra sequence called a “toehold” is attached to the end of the DNA strand. Using this toehold, the DNA molecular machines are operated by adding and removing specific DNA strands for complex movements. When a DNA strand fully complementary to a toehold‐containing DNA is added, the initial complementary strand without the toehold is selectively removed by strand displacement [16]. The thermodynamic stabilization energy for hybridization works as “fuel” to provide the mechanical motion of the DNA molecular machine. Using this strategy, DNA tweezers that perform open–close motions were constructed ( Figure 1.2d) [8]. Seeman and coworkers created a molecular machine combining DNA nanostructures. Using the helical rotation of dsDNA during the B–Z transition, in which the dsDNA conformation changes from a right‐handed (B‐form DNA) to a left‐handed (Z‐form DNA) conformation, a reciprocating motion of the DNA nanostructure was observed [17]. In addition, they developed molecular machines that perform 180° rotation at the ends of two adjacent dsDNAs, called PX‐JX 2devices, by hybridization and removal of DNA strands ( Figure 1.2e) [9]. Both the PX and JX 2states were directly observed by atomic force microscopy (AFM). These dynamic systems were also introduced to DNA origami to operate DNA nanodevices (see Section 1.9).
1.2 Two‐Dimensional DNA Origami
DNA origami has enabled the construction of a wide variety of 2D structures approximately 100 nm in size, including rectangles, triangles, and even a smiley face and five‐pointed star ( Figure 1.3) [6]. In this method, a long single‐stranded DNA (M13mp18; 7249 nucleotides) and sequence‐designed complementary strands (called “staple strands”; most of which are 32‐mer) are mixed and then annealed from 95 °C to room temperature over two hours, resulting in the formation of target structures by self‐assembly ( Figure 1.3a). The structure can be imaged by AFM, and the assembled structure formed according to a design. To create 2D DNA origami structures, adjacent dsDNAs should be connected to each other via a crossover. In this design, the geometry of the double helices involved has three helical rotations for 32 base pairs ( Figure 1.3b). For example, two neighboring crossovers of the central dsDNA in an arrangement of three adjacent dsDNAs should be located at the opposite sites (rotated at 180°, 0.5 turns); therefore, the crossovers should be separated by 16 base pairs (1.5 turns). This rule should be preserved to maintain stable planar structure when placing multiple staple strands on the scaffold. DNA origami structures are formed using many different staple strands, so DNA hairpins can be placed as markers at any position on the surface of the DNA structure. A hairpin DNA (dumbbell‐type) used as a topological marker was observed as a dot by AFM imaging ( Figure 1.3c). In this case, hairpins are placed perpendicular to the surface of the origami; therefore, each hairpin should be placed at a position eight base pairs from the crossover (270° rotation). The distance between the centers of the adjacent staples is approximately 6 nm, so the adjacent hairpins can be observed as different spots according to the spatial resolution of AFM. Using the hairpin markers, patterns, such as the map of a hemisphere ( Figure 1.3c), can be displayed precisely on the DNA origami surface.
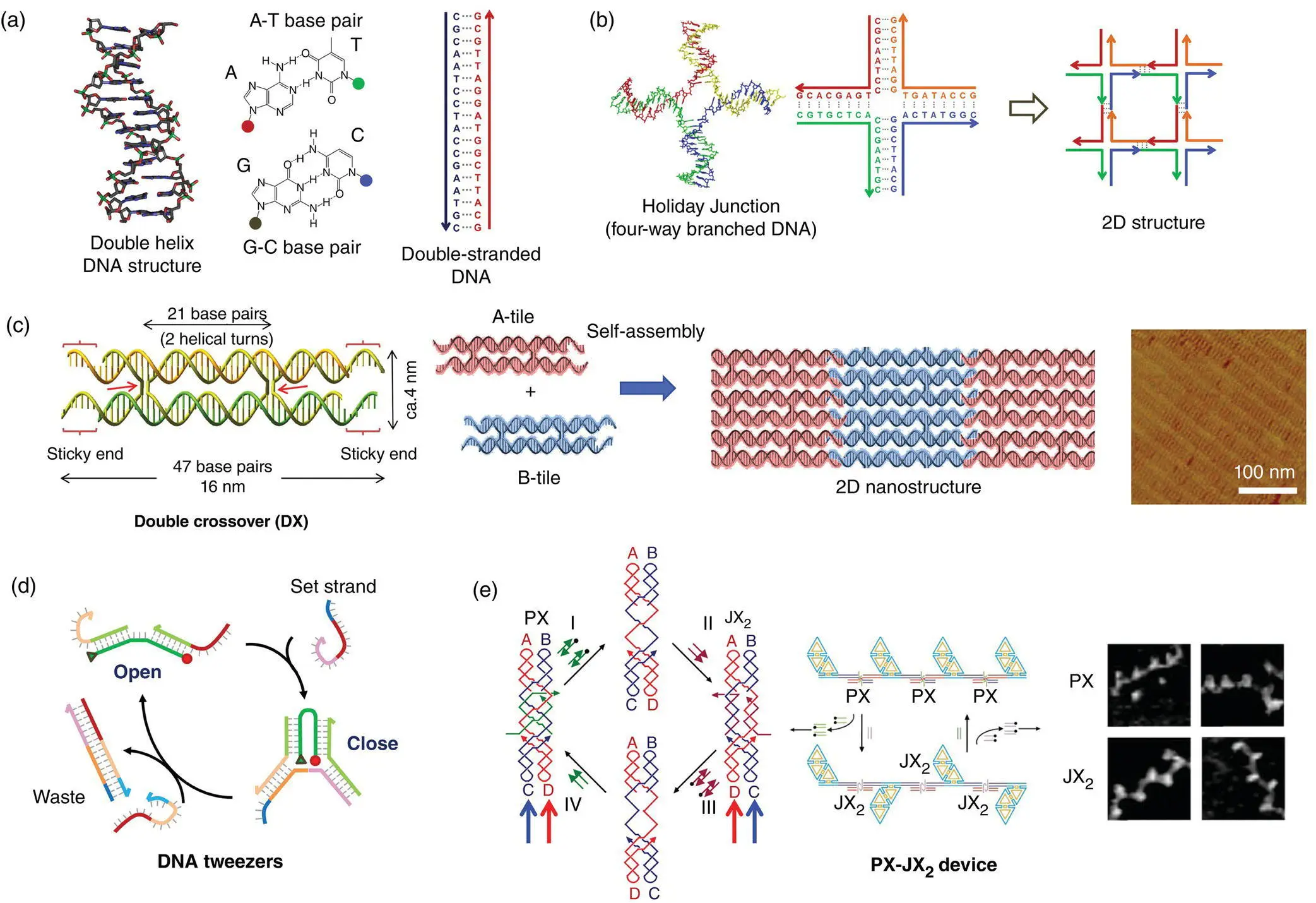
Figure 1.2 DNA nanotechnology before the emergence of DNA origami. (a) DNA double helix structure, base pair, and double‐stranded DNA (dsDNA). (b) Holliday junction structure, four‐way junction, and conceptual diagram for construction of 2D structure.
Source: Modified from Seeman [1].
(c) Double crossover structure, in which two dsDNAs are connected by four‐way branched strands (crossover; arrows). Two‐dimensional periodic structure was formed by self‐assembly using two double‐crossover components (A‐tile and B‐tile* with hairpin) with sticky ends (complementary single‐stranded DNAs at the ends). AFM image of the self‐assembled 2D nanostructure.
Source: Modified from Winfree et al. [10]
(d) Dynamic open/close behavior of DNA tweezers operated by strand displacement using toehold containing DNA strands.
Source: Modified from Yurke et al. [8].
(e) PX‐JX 2device to exchange the bottom part of by insertion and removal of the strands. The structures can be observed in AFM images.
Source: Yan et al. [9]/with permission of Springer Nature.
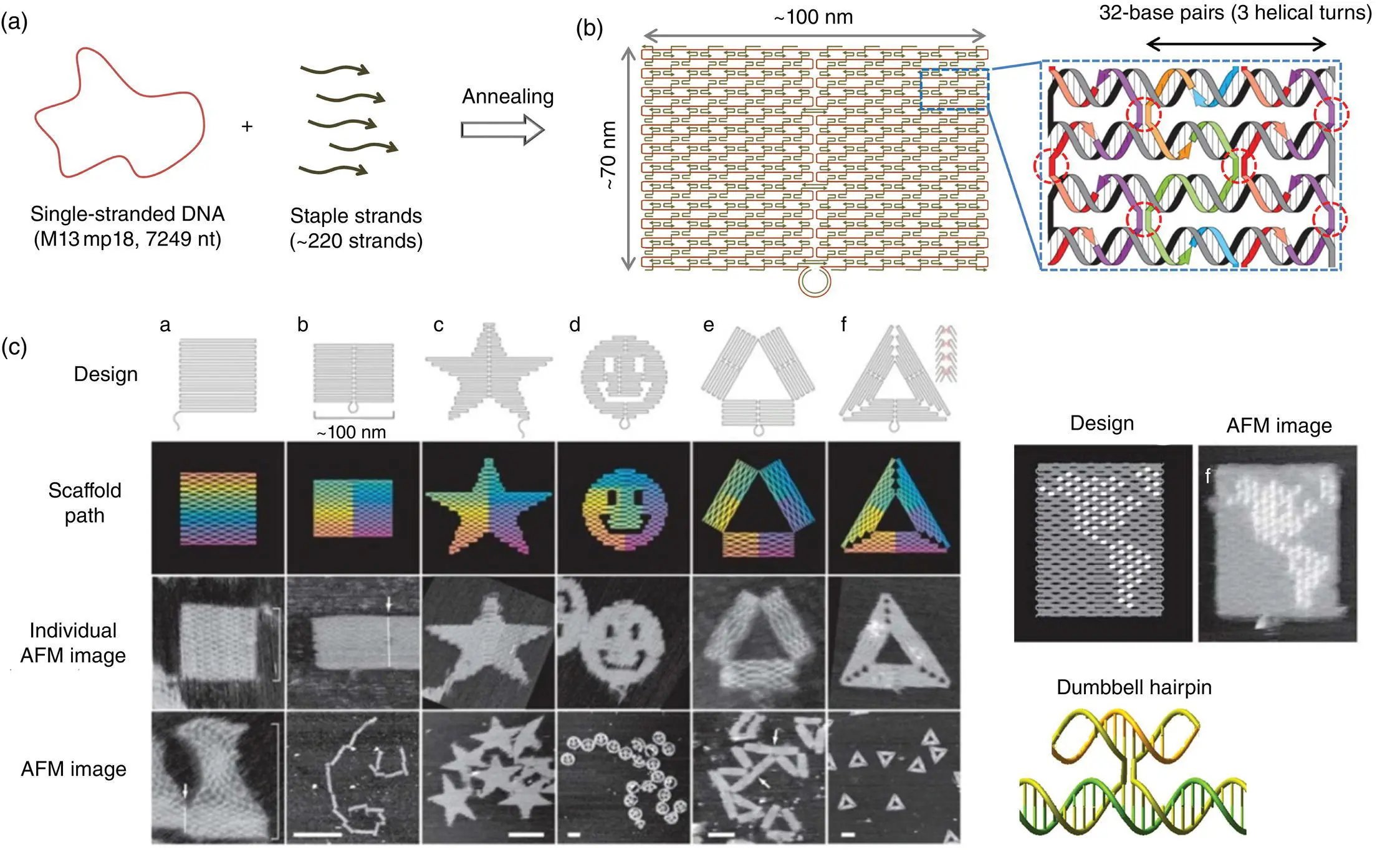
Figure 1.3 DNA origami. (a) Method to prepare a DNA origami structure from the template single‐stranded DNA and staple strands. (b) Design of a self‐assembled DNA origami structure and geometry of the incorporated double‐stranded DNAs. Colored strands and a gray/black strand represent staple strands and template single‐stranded DNA, respectively. Staple strands connect the adjacent duplexes with crossovers. Inset: Structure of hairpin DNA for a topological marker. (c) Design and AFM images of self‐assembled DNA origami structures. Drawing of a hemisphere on the DNA origami with hairpin DNAs (white dots) and an AFM image of the assembled DNA origami.
Source: Rothemund [6]/with permission of Springer Nature.
1.3 Programmed Arrangement of Multiple DNA Origami Components
The programmed arrangement of multiple DNA origami structures is an important technique for preparing larger structures, particularly in terms of integrating complex functions. We explored techniques for arranging multiple DNA origami components and developed methods to arrange rectangular DNA origami tiles horizontally in a programmed fashion [18]. Because the ends of the helical axes align at both edges of the DNA origami rectangles, origami tiles horizontally assemble via π‐interactions at the edges in a predictable fashion [18]. Specific concave and convex connectors were introduced into DNA origami tiles to precisely align neighboring tiles in a shape‐fitting manner. DNA tiles could be correctly assembled by shape and sequence complementarity, where the complementary strands were introduced into the concavity and the convex connectors. After self‐assembly of three, four, and five tiles, the DNA tiles were aligned and oriented in the same direction in a designed manner. For identification of the DNA tiles, hairpin markers were introduced onto individual tiles. After self‐assembly, judging from the order of the markers, the five tiles were aligned correctly. This method was further expanded vertically to form 2D assemblies.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «DNA Origami»
Представляем Вашему вниманию похожие книги на «DNA Origami» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «DNA Origami» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












