Stephen R. Bolsover - Cell Biology
Здесь есть возможность читать онлайн «Stephen R. Bolsover - Cell Biology» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Cell Biology
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Cell Biology: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Cell Biology»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
Cell Biology: A Short Course
Cell Biology: A Short Course
Cell Biology: A Short Course
Cell Biology — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Cell Biology», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Single‐Stranded DNA‐Binding Proteins
As soon as DnaB unwinds the two parental strands, they are engulfed by single‐stranded DNA‐binding proteins. These proteins bind to adjacent groups of 32 nucleotides. DNA covered by single‐stranded DNA‐binding proteins is rigid, without bends or kinks. It is, therefore, an excellent template for DNA synthesis. Single‐stranded binding proteins are sometimes called helix‐destabilizing proteins.
 BIOCHEMISTRY OF DNA REPLICATION
BIOCHEMISTRY OF DNA REPLICATION
In prokaryotes the synthesis of a new DNA molecule is catalyzed by the enzyme DNA polymerase III.Its substrates are the four deoxyribonucleoside triphosphates, dGTP, dATP, dTTP, and dCTP. DNA polymerase III catalyzes the formation of a phosphodiester bond ( Figure 3.3on page 37) between the 3′ hydroxyl of the sugar residue on the most recently added nucleotide and the 5′ phosphate of the incoming nucleotide. The elongation of the new DNA molecule takes place in the 5′ to 3′ direction ( Figure 4.2a). The base sequence of a newly synthesized DNA strand is dictated by the base sequence of its parental strand. If the sequence of the template strand is 3′ CATCGA 5′, then that of the daughter strand is 5′ GTAGCT 3′. In eukaryotes, DNA replication is performed by three isoforms, DNA polymerases α, δ, and ε, but the mechanism is much the same.
DNA polymerase III can only add a nucleotide to a free 3′‐hydroxyl group and therefore synthesizes DNA in the 5′ to 3′ direction. The template strand is read in the 3′ to 5′ direction. However, the two strands of the double helix are antiparallel. They cannot be synthesized in the same direction because only one has a free 3′‐hydroxyl group, the other has a free 5′‐phosphate group. No DNA polymerase has been found that can synthesize DNA in the 3′ to 5′ direction, that is, by attaching a nucleotide to a 5′ phosphate, so the synthesis of the two daughter strands must differ. One strand, the leading strand,is synthesized continuously while the other, the lagging strand,is synthesized discontinuously. DNA polymerase III can synthesize both daughter strands, but must make the lagging strand as a series of short 5′ to 3′ sections ( Figure 4.1). These fragments of DNA, called Okazaki fragmentsafter Reiji Okazaki who discovered them in 1968, are then joined together by DNA ligase.
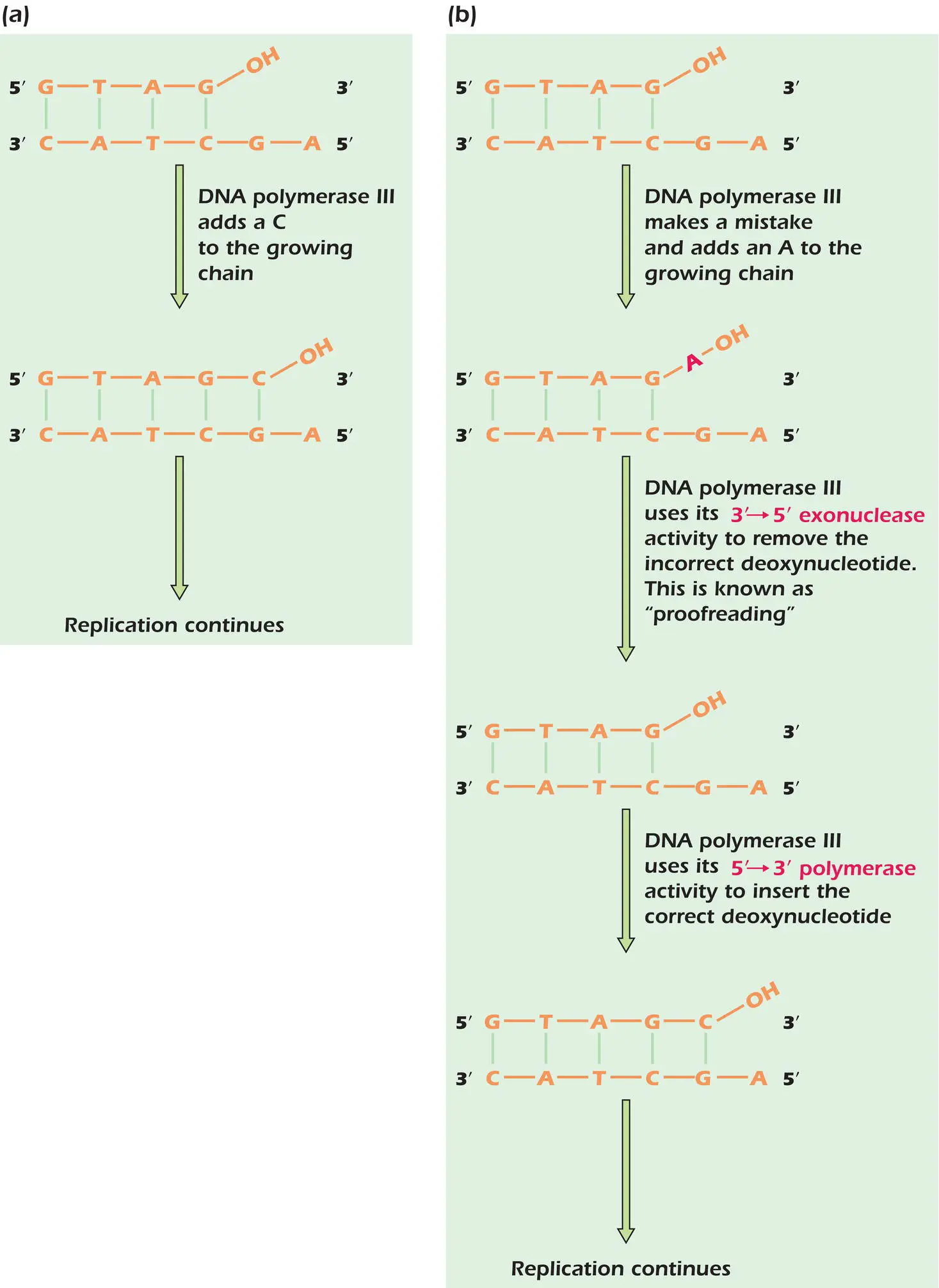
 Figure 4.2.DNA polymerase III can correct its own mistakes.
Figure 4.2.DNA polymerase III can correct its own mistakes.
Medical Relevance 4.1 Inhibiting DNA Polymerase Fights Cancer
Drugs that inhibit DNA replication prevent cells dividing. Cytarabine (also known as cytosine arabinoside and arabinofuranosyl cytidine) is sold as Cytosar‐U, Tarabine PFS, Depocyt, and AraC. It inhibits eukaryote DNA polymerases α, δ, and ε, and is therefore widely used to treat cancers. It is on the World Health Organization's model list of essential medicines.
Example 4.1 The Meselson and Stahl Experiment
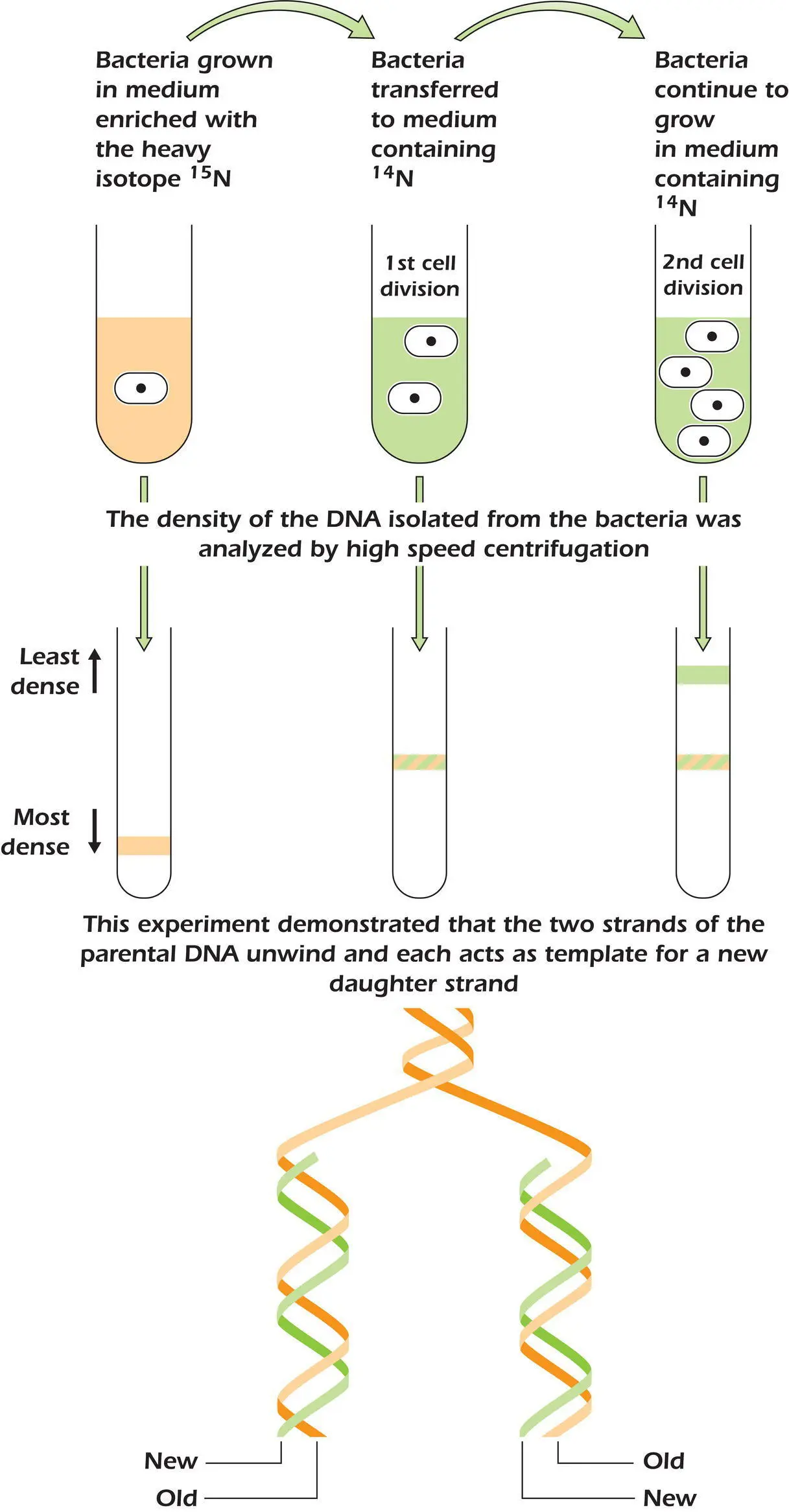
In 1958 Matthew Meselson and Franklin Stahl designed an ingenious experiment to test whether each strand of the double helix does indeed act as a template for the synthesis of a new strand. They grew the bacterium E. coli in a medium containing the heavy isotope 15N that could be incorporated into new DNA molecules. After several cell divisions they transferred the bacteria, now containing “heavy” DNA, to a medium containing only the lighter, normal, isotope 14N. Any newly synthesized DNA molecules would therefore be lighter than the original parent DNA molecules containing 15N. The difference in density between the heavy and light DNAs allows their separation using very high‐speed centrifugation. The results of this experiment are illustrated in the figure. DNA isolated from cells grown in the 15N medium had the highest density and migrated the furthest during centrifugation. The lightest DNA was found in cells grown in the 14N medium for two generations, whereas DNA from bacteria grown for only one generation in the lighter 14N medium had a density halfway between these two. This is exactly the pattern expected if each strand of the double helix acts as a template for the synthesis of a new strand. The two heavy parental strands separated during replication, with each acting as a template for a newly synthesized light strand, which remained bound to the heavy strand in a double helix. The resulting DNA was therefore of intermediate density. Only in the second round of DNA replication, when the light strands created during the first round of replication were allowed to act as templates for the construction of complementary light strands, did DNA double helices composed entirely of 14N‐containing building blocks appear.
DNA Synthesis Requires an RNA Primer
DNA polymerase III cannot itself initiate the synthesis of DNA. The enzyme primaseis needed to catalyze the formation of a short stretch of RNA complementary in sequence to the DNA template strand ( Figure 4.1). This RNA chain, the primer, is needed to prime (or start) the synthesis of the new DNA strand. DNA polymerase III catalyzes the formation of a phosphodiester bond between the 3′‐hydroxyl group of the RNA primer and the 5′‐phosphate group of the appropriate deoxyribonucleotide. Several RNA primers are made along the length of the lagging strand template. Each is extended in the 5′ to 3′ direction by DNA polymerase III until it reaches the 5′ end of the next RNA primer. In prokaryotes the lagging strand is primed about every 1000 nucleotides whereas in eukaryotes this takes place every 200 nucleotides.
RNA Primers Are Removed
Once the synthesis of the DNA fragment is complete, the RNA primers must be replaced by deoxyribonucleotides. In prokaryotes the enzyme DNA polymerase I removes ribonucleotides using its 5′ to 3′ exonucleaseactivity and then uses its 5′ to 3′ polymerizing activity to incorporate deoxyribonucleotides. In this way, the entire RNA primer is replaced by DNA. Synthesis of the lagging strand is completed by the enzyme DNA ligase, which joins the DNA fragments together by catalyzing the formation of phosphodiester bonds between adjacent fragments.
Eukaryotic organisms use an enzyme called ribonuclease Hto remove their RNA primers. These enzymes break phosphodiester bonds in an RNA strand that is hydrogen‐bonded to a DNA strand. Within each cell, ribonuclease H2 is involved in removing RNA primers during replication of nuclear genomic DNA and ribonuclease H1 is involved in removing RNA primers during replication of mitochondrial genomic DNA.
The Self‐Correcting DNA Polymerase
The genome of E. coli consists of about 4.6 × 10 6base pairs of DNA. DNA polymerase III makes a mistake about every 1 in 10 5bases and joins an incorrect deoxyribonucleotide to the growing chain. If unchecked, these mistakes would lead to a catastrophic mutation rate. Fortunately, DNA polymerase III has a built‐in proofreading mechanism that corrects its own errors. If an incorrect base is inserted into the newly synthesized daughter strand, the enzyme recognizes the change in shape of the double‐stranded molecule, which arises through incorrect base pairing, and DNA synthesis stops ( Figure 4.2b). DNA polymerase III then uses its 3′ to 5′ exonuclease activity to remove the incorrect deoxyribonucleotide and replace it with the correct one. DNA synthesis then continues. DNA polymerase III hence functions as a self‐correcting enzyme.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Cell Biology»
Представляем Вашему вниманию похожие книги на «Cell Biology» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Cell Biology» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












