Genotyping by Sequencing for Crop Improvement
Здесь есть возможность читать онлайн «Genotyping by Sequencing for Crop Improvement» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Genotyping by Sequencing for Crop Improvement
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:4 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 80
- 1
- 2
- 3
- 4
- 5
Genotyping by Sequencing for Crop Improvement: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Genotyping by Sequencing for Crop Improvement»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
A thoroughly up-to-date exploration of genotyping-by-sequencing technologies and related methods in plant science Genotyping by Sequencing for Crop Improvement,
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement
Genotyping by Sequencing for Crop Improvement — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Genotyping by Sequencing for Crop Improvement», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
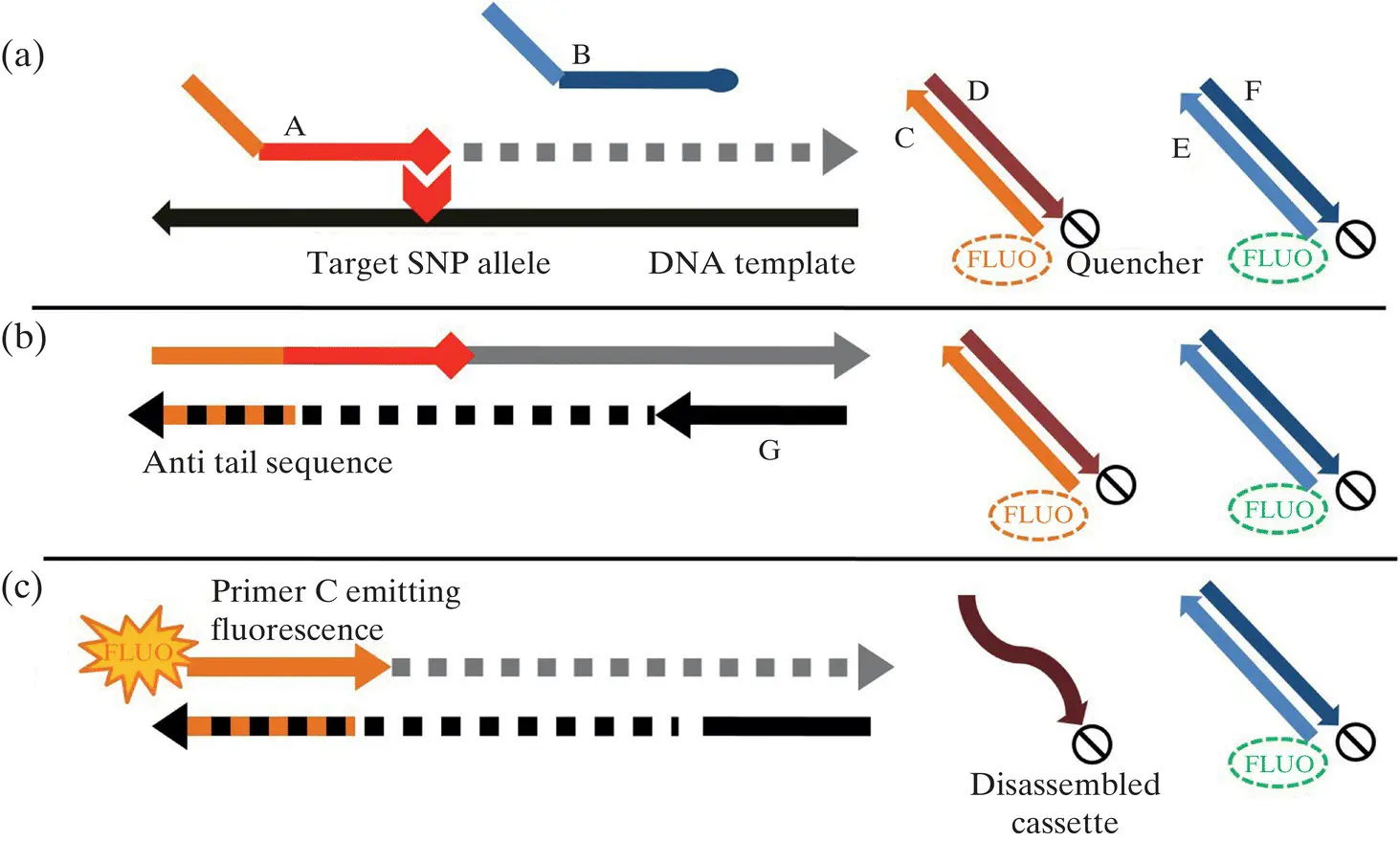
Figure 1.2 Steps in KASP reaction: (a) annealing: allele‐specific primer binds to target SNP, (b) extension: anti tail sequence generation leading to disassembly of allele‐specific FRET cassette, and (c) fluorescent emission: sample emitting fluorescence on exposing to a specific wavelength.
Source: The figure is reproduced from Rosas et al. (2014) available with CC‐BY‐4.0.
1.6 SNP Databases
SNP databases correspond to a publically available archive of genetic information of economically important species. Recent developments in genome sequencing technologies have ushered the era of cost‐effective, high‐throughput genomics resulting in the creation of huge datasets of sequence information. In a similar manner, developments in computing facilities and data sciences had enabled us to compare, categorize, and compute relationship matrices among and between species leading to the creation of databases. As numerous labs are working on the same species, collaborative consortiums were established to avoid redundancy leading to high‐quality SNP databases containing a range of molecular variations constituting SNP’s, insertions, and deletions (InDels), trait‐specific characterized SNP’s and called variants. Table 1.3enlists some important SNP databases corresponding to humans, model genetic organisms, and important crop species. These public databases serve as an important resource for crop improvement for genetic diversity analysis, establishing a genetic association and linkage disequilibrium studies.
Table 1.3 List of important online SNP databases.
| SNP database | Organism | URL |
|---|---|---|
| dbSNP | Human | http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp |
| Ensembl | Human | http://www.ensembl.org/Homo_sapiens/index.html |
| 1001 Genomes | Arabidopsis | https://1001genomes.org/ |
| CropSNPdb | Brassica crops, wheat | http://snpdb.appliedbioinformatics.com.au |
| SNP‐Seek Database | Rice 3K panel | https://snp‐seek.irri.org/ |
| MaizeSNPDB | 1210 Maize inbred lines | http://150.109.59.144:3838/MaizeSNPDB/ |
| CerealDB | Wheat | http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/indexNEW.php |
1.7 Application of Molecular Markers
1.7.1 Application of Molecular Markers in Crop Improvement
Molecular markers have several applications in genetic studies and crop improvement programs. These have been used in the development of saturated linkage maps, gene/QTL mapping, map‐based cloning of genes, orthologous gene mapping, and marker‐assisted transfer of targeted genes/QTLs in the background of different cultivars/lines. Saturation of linkage maps refers to increased marker density to cover the entire chromosomal region. In general, when molecular markers are arranged on a linkage map with less than 1 cM distance apart, is considered as saturated linkage map. The development of saturated linkage maps could only be possible with the availability of molecular markers. These maps are prerequisite for gene/QTL mapping, map‐based cloning of genes, and MAS. Several molecular markers‐based saturated linkage maps have been developed in crop plants including rice (Harushima et al. 1998; McCouch et al. 2002; IRGSP 2005; Zhu et al. 2017; Kumar et al. 2018), wheat (Somers et al. 2004; Song et al. 2005; Poland et al. 2012; Li et al. 2015; Hussain et al. 2017), maize (Sharopova et al. 2002; Zhou et al. 2016; Su et al. 2017), and tomato (Tanksley et al. 1992; Haanstra et al. 1999; Sim et al. 2012). In one of the studies, a rice genetic map helped to enrich the genetic region of Ph1 locus of wheat and facilitated the identification of candidate genes governing the locus (Sidhu et al. 2008).
Plant breeders have relied heavily on generating new gene combinations and selecting these new gene combinations empirically. Though phenotype‐based selection has largely been successful, MAS has improved the efficiency and precision of selection. MAS can be practiced more efficiently for characters whose phenotypic selection is difficult. As an example, selecting a fertility restorer gene in segregating generations need test crossing before subsequent backcrossing. However, if such genes are tagged with molecular markers, desirable plants with fertility restorer genes (in heterozygous condition), can be identified and backcrossed. Similarly, if desirable genes conferring tolerance to abiotic stresses are tagged, these can be selected easily in segregating generations. Also, genetic markers can be assayed in nontarget areas such as growth chambers, greenhouses, or off‐season nurseries, thus permitting more rapid progress. The efficiencies of scale and time accorded by DNA markers are valuable in breeding horticultural plants where fewer individuals might save several hectares and fewer generations may save several decades (Paterson et al. 1991). MAS has been used efficiently in (i) gene pyramiding (Huang et al. 1997; Singh et al. 2001; Bhatia et al. 2011; Kumar et al. 2013; Yasuda et al. 2015), (ii) marker‐assisted alien introgressions (Jena et al. 1992; Brar and Dhaliwal 1997; Elkot et al. 2015), and (iii) simultaneous identification and pyramiding of QTLs from primitive cultivars and wild species (Tanksley and Nelson 1996; Tanksley et al. 1996; Fulton et al. 1997; Xiao et al. 1996, 1998).
1.7.2 Role of Molecular Markers in Germplasm Characterization
Molecular markers are also used in DNA fingerprinting for varietal identification, germplasm evaluation, phylogenetic and evolutionary studies, etc. The molecular marker‐based DNA fingerprinting data are useful for the characterization of plant germplasm accessions, quantification of genetic diversity, and protection of proprietary germplasm (Smith and Smith 1992). Molecular markers have been utilized to distinguish closely related crop cultivars (Melchinger et al. 1991; Paull et al. 1998), in sex identification of dioecious plants (Parasnis et al. 1999). They are also used to understand evolutionary relationships within and between species, genera, or higher taxonomic groups. Such studies involve large number of markers to study similarities and differences among taxa (Paterson et al. 1991). Although phylogeny has been established for many plant species based on morphological markers, biochemical markers, and chromosome homology, the genetic markers have enhanced our understanding of phylogeny. In one important study, DNA‐based markers enabled the designation of GG for Oryza granulata and HHJJ for Oryza ridleyi ( Aggarwal et al. 1997 ) .
1.7.3 Deployment of Molecular Markers in Plant Variety Protection and Registration
The current system of plant variety protection and registration using assessment of DUS (Distinctness, Uniformity, and Stability) characteristics predominantly relies upon morphological traits which are quite laborious, time‐consuming, requires skills, expertise, and evaluation under special designs for most quantitative traits. With the advent of novel breeding technologies, new varieties differ only for few traits or at few loci which make the process of detecting distinctness in varieties, a challenging task. Even with increasing numbers of plant varieties, DUS testing is becoming quite expensive. In a review by Jamali et al. (2019), DNA‐based molecular markers have been proposed to be a reliable alternative for conducting DUS testing. Molecular markers not only cut cost, time and labor but will also help in the proper sharing of Plant Breeder’s Rights with an assessment of few distinct traits, particularly in essentially derived varieties.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Genotyping by Sequencing for Crop Improvement»
Представляем Вашему вниманию похожие книги на «Genotyping by Sequencing for Crop Improvement» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Genotyping by Sequencing for Crop Improvement» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












