Martinez J. Hewlett - Basic Virology
Здесь есть возможность читать онлайн «Martinez J. Hewlett - Basic Virology» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Basic Virology
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Basic Virology: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Basic Virology»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
The basics of virology Virological techniques Molecular biology Pathogenesis of human viral disease The 4th edition includes new information on the SARS, MERS and COVID-19 coronaviruses, hepatitis C virus, influenza virus, as well as HIV and Ebola. New virological techniques including bioinformatics and advances in viral therapies for human disease are also explored in-depth. The book also includes entirely new sections on metapneumoviruses, dengue virus, and the chikungunya virus.
Basic Virology — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Basic Virology», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
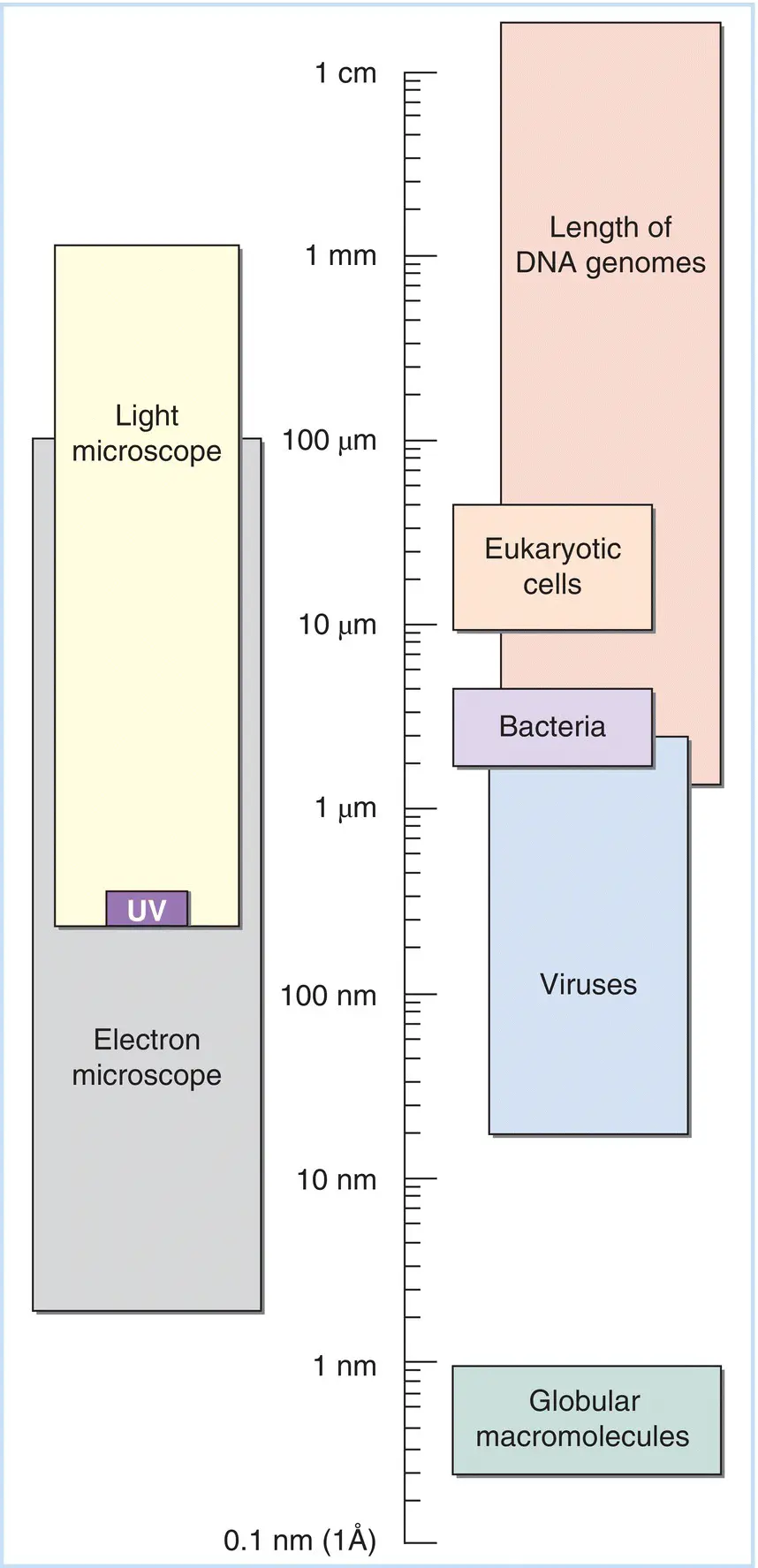
Figure 5.1A scale of dimensions for biologists. The wavelength of a photon or other subatomic particle is a measure of its energy and its resolving power. An object with dimensions smaller than the wavelength of a photon cannot interact with it, and thus is invisible to it. The dimensions of some important biological features of the natural world are shown. Note that the wavelength of ultraviolet (UV) light is between 400 and 280 nm; objects smaller than that, such as viruses and macromolecules, cannot be seen in visible or UV light. The electron microscope can accelerate electrons to high energies; the resulting short wavelengths can resolve viruses and biological molecules. Note that the length of DNA is a measure of its information content, but since DNA is essentially “one‐dimensional,” it cannot be resolved by light.
The development of self‐consistent classification schemes for this plethora of entities is a major challenge for virologists. Good classification schemes have a major role in helping organize the growing flood of detailed genetic and molecular information concerning viruses and their genes. Further, a valid classification scheme provides an important framework for understanding the different ways that viruses can utilize cellular and their own genes in maintaining themselves within the biosphere. Finally, valid classifications provide useful guides to our understanding of the origins of various virus groups, and the relationships between viruses in the same group and divergent groups.
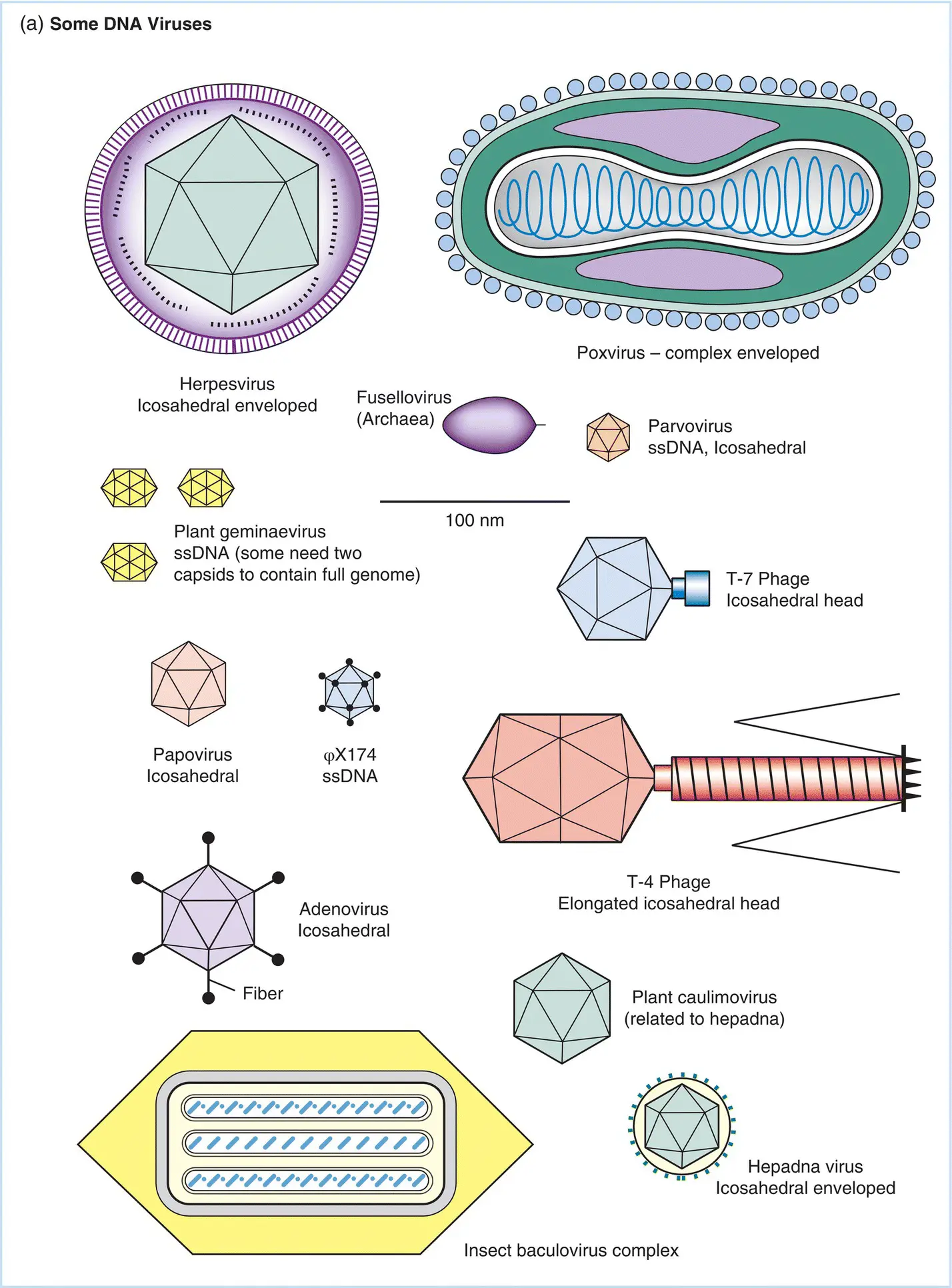
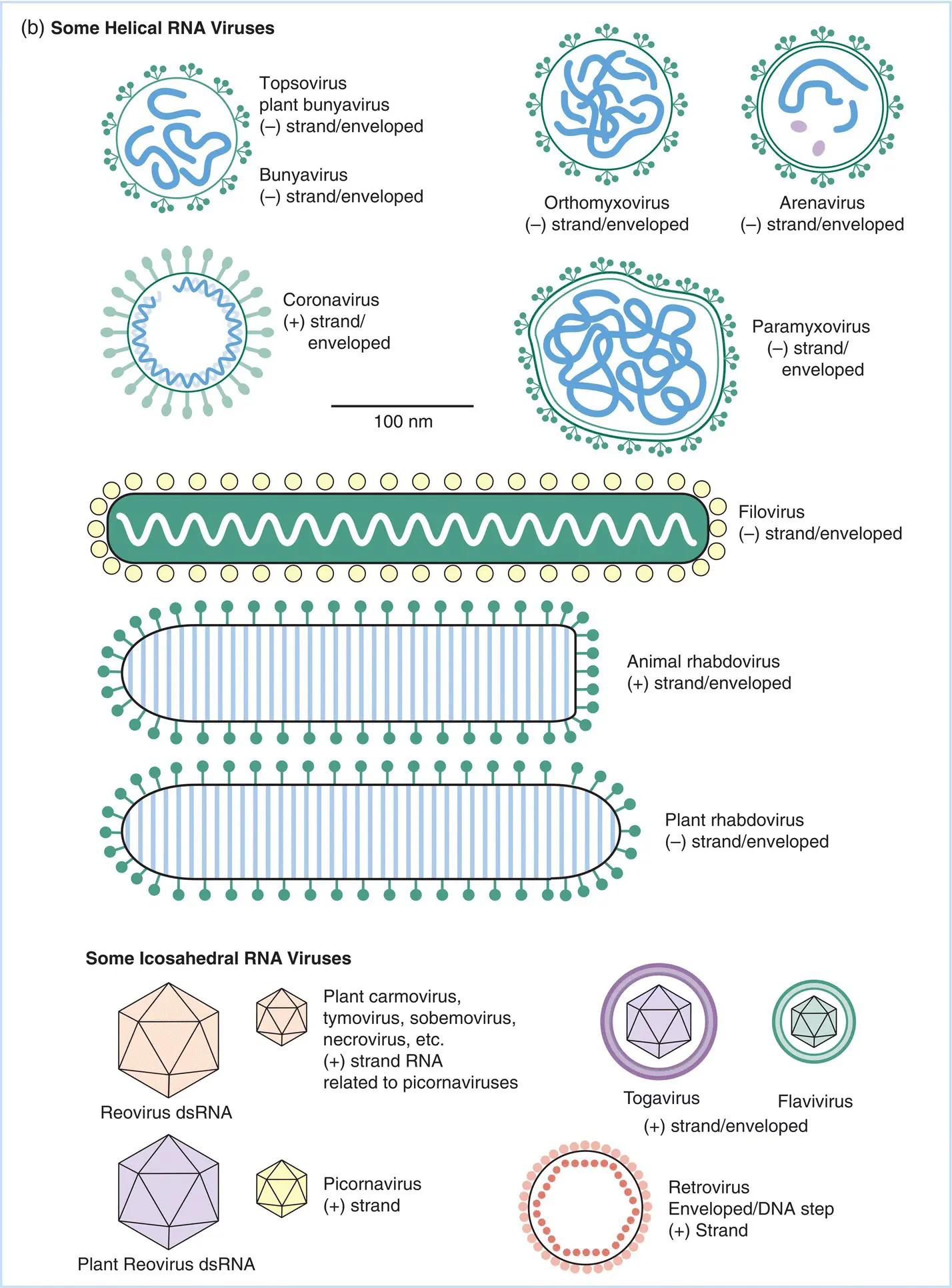
Figure 5.2The structure and relative sizes of a number of (a) DNA and (b) RNA viruses. The largest viruses shown have dimensions approaching 300–400 nm and can be just resolved as refractile points in a high‐quality ultraviolet‐light microscope. The smallest dimensions of viruses shown here are on the order of 25 nm. Classifications of viruses based on the type of nucleic acid serving as the genome and the shape of the capsid are described in the text. ss: Single stranded; ds: double stranded.
The International Committee on Taxonomy of Viruses (ICTV) was created at an international conference on microbiology in Moscow in 1966 in order to develop a single, universal taxonomic scheme for all the viruses infecting animals (vertebrates, invertebrates, and protozoa), plants (higher plants and algae), fungi, bacteria, and archaea. Its membership is made up of distinguished virologists throughout the world, and it has issued periodic reports describing its progress, and its problems, as well as databases containing the properties of viruses and appropriate computer‐based tools for utilizing such databases. One of the notable achievements of this group and the community of virologists that it represents is the recognition of a limited number of viral features that can be used for classification; these include the nature of the viral genome, the presence of an envelope, and the morphology of the virus particle. The classification scheme uses the designation of “family,” even though these phylogenetic terms do not strictly apply in the case of viruses. Table 5.1lists the families of this scheme, in alphabetical order, as of their 2018 report.
Table 5.1Classification of viruses according to the ICTV.
| Family | Nature of the genome | Presence of an envelope | Morphology | Genome configuration | Genome size (kbp or kb) | Host |
|---|---|---|---|---|---|---|
| Abyssoviridae | ssRNA | + | ||||
| Ackermannviridae | dsDNA | − | Icosahedral tailed | 1 linear | 155 | Bacteria |
| Adenoviridae | dsDNA | − | Isometric | 1 linear | 28–45 | Vertebrates |
| Alloherpesviridae | dsDNA | + | Isometric | 1 linear | 134–248 | Vertebrates |
| Alphaflexiviridae | ssRNA | − | Filamentous | 1 + segment | 7–9 | Plants |
| Alphasatellitidae | ssDNA | − | N/A | 1 linear | 1.3–1.4 | Plants |
| Alphatetraviridae | ssRNA | − | Isometric | 1 linear | 6.5 | Invertebrates |
| Alvernaviridae | ssRNA | − | Isometric | 1 linear | 4.4 | Dinoflagellates |
| Amalgaviridae | dsRNA | − | Isometric | 1 linear | 3.5 | Plants |
| Ampullaviridae | dsDNA | + | Bottle‐shaped | 1 linear | 23.8 | Archaea |
| Anelloviridae | ssDNA | − | Isometric | 1 linear | 3–4 | Vertebrates |
| Arenaviridae | NssRNA | + | Spherical | 2 ± segments | 11 | Vertebrates |
| Arteriviridae | ssRNA | + | Isometric | 1 + segment | 13–16 | Vertebrates |
| Artoviridae | ssRNA | + | Spherical | 1 − linear | 12.3 | Invertebrates and vertebrates |
| Ascoviridae | dsDNA | + | Reniform | 1 linear | 100–180 | Invertebrates |
| Asfarviridae | dsDNA | + | Spherical | 1 circular | 170–190 | Vertebrates |
| Aspiviridae | ssRNA | − | Filamentous | 3–4 – linear | 11.3–12.5 | Plants |
| Astroviridae | ssRNA | − | Isometric | 1 + segment | 7–8 | Vertebrates |
| Avsunviroidae | ssRNA | N/A | N/A | 1 + circular | 0.25 | Plant viroids |
| Bacilladnaviridae | ssDNA | − | Isometric | 1 circular | 5–6 | Diatoms |
| Baculoviridae | dsDNA | + | Bacilliform | 1 circular supercoiled | 80–180 | Invertebrates |
| Barnaviridae | ssRNA | − | Bacilliform | 1 + segment | 4 | Fungi |
| Belpaoviridae | ssRNA (RT) | N/A | N/A | 1 linear | ? | Invertebrates |
| Benyviridae | ssRNA | − | Rod‐shaped | 4/5 + segments | 14–16 | Plants |
| Betaflexiviridae | ssRNA | − | Filamentous | 1 linear | 6.5–9 | Plants, fungi |
| Bicaudaviridae | dsDNA | + | Lemon‐shaped | 1 circular | 62 | Archaea |
| Bidnaviridae | ssDNA | − | Isometric | 2 linear | 6 and 6.5 | Invertebrates |
| Birnaviridae | dsRNA | − | Isometric | 2 segments | 6 | Vertebrates and invertebrates |
| Bornaviridae | NssRNA | + | Spherical | 1 − segment | 6 | Vertebrates |
| Bromoviridae | ssRNA | − | Isometric | 3 + segments | 8–9 | Plants |
| Caliciviridae | ssRNA | − | Isometric | 1 + segment | 7–8 | Vertebrates |
| Carmotetraviridae | ssRNA | − | Isometric | 1 linear | 6.1 | Invertebrates |
| Caulimoviridae | dsDNA‐RT | − | Isometric, bacilliform | 1 circular | 8 | Plants |
| Chrysoviridae | dsRNA | − | Isometric | 4 linear | 2.9–3.6 | Fungi |
| Chuviridae | ssRNA | ? | ? | ? (negative sense) | ? | Invertebrates |
| Circoviridae | ssDNA | − | Isometric | 1 circular | 2 | Vertebrates |
| Clavaviridae | dsDNA | Bacilliform | 1 circular | 5.3 | Archaea | |
| Closteroviridae | ssRNA | − | Filamentous | 1/2 + segments | 15–19 | Plants |
| Coronaviridae | ssRNA | + | Isometric | 1 + segment | 27–31 | Vertebrates |
| Corticoviridae | dsDNA | − | Isometric | 1 circular supercoiled | 9 | Bacteria |
| Cruliviridae | ssRNA | ? | ? | ? (negative sense) | ? | Invertebrates |
| Cystoviridae | dsRNA | + | Spherical | 3 segments | 13 | Bacteria |
| Deltaflexiviridae | ssRNA | ? | ? | 1 + sense | 6–8 | Fungi, plants |
| Dicistroviridase | ssRNA | − | Isometric | 1 linear | 8.5–10.2 | Invertebrates |
| Endornaviridae | dsRNA | N/A | No true capsid | 1 linear | 14 | Plants |
| Euroniviridae | ssRNA | ? | ? | ? | ? | ? |
| Filoviridae | NssRNA | + | Bacilliform | 1 − segment | 19 | Vertebrates |
| Fimoviridae | ssRNA | + | Spherical | 4 − sense segments | 12 | Plants |
| Flaviviridae | ssRNA | + | Isometric | 1 + segment | 10–12 | Vertebrates |
| Fuselloviridae | dsDNA | + | Lemon‐shaped | 1 circular supercoiled | 15 | Archaea |
| Gammaflexiviridae | ssRNA | − | Filamentous | 1 linear | 6.8 | Plants |
| Geminiviridae | ssDNA | − | Isometric | 1 or 2 circular | 3–6 | Plants |
| Genomoviridae | ssDNA | − | Isometric | 1 circular +/− | 2.17 | Mammals, birds, fungi |
| Globuloviridae | dsDNA | + | Spherical | 1 circular | 20–30 | Archaea |
| Guttaviridae | dsDNA | + | Ovoid | 1 circular | 20 | Archaea |
| Hantaviridae | ssRNA | + | Spherical | 3 linear negative sense | 11–20 | Humans, rodents |
| Hepadnaviridae | dsDNA‐RT | + | Spherical | 1 circular | 3 | Vertebrates |
| Hepeviridae | ssRNA | − | Isometric | 1 linear | 7.2 | Vertebrates |
| Herpesviridae | dsDNA | + | Isometric | 1 linear | 125–240 | Vertebrates |
| Hypoviridae | dsRNA | − | Pleomorphic | 1 segment | 12 | Fungi |
| Hytrosaviridae | dsDNA | + | Filamentous | 1 circular | 120–190 | Insects |
| Iflaviridae | ssRNA | − | Isometric | 1 linear | 8.8–9.7 | Invertebrates |
| Inoviridae | ssDNA | − | Filamentous | 1 + circular | 7–9 | Bacteria, mycoplasmas |
| Iridoviridae | dsDNA | − | Isometric | 1 linear | 140–383 | Vertebrates and invertebrates |
| Lavidaviridae | dsDNA | − | Isometric | 1 circular | 17–30 | Protists |
| Leviviridae | ssRNA | − | Isometric | 1 + segment | 3–4 | Bacteria |
| Lipothrixviridae | dsDNA | + | Rod‐shaped | 1 linear | 16 | Archaea |
| Lispiviridae | ssRNA | + (?) | Spherical (?) | 1, linear | 12 | Arachnids |
| Luteoviridae | ssRNA | − | Isometric | 1 + segment | 6 | Plants |
| Malacoherpesviridae | dsDNA | + | Spherical | 1 linear | 150 | Mollusks |
| Marnaviridae | ssRNA | − | Isometric | 1 linear | 8.6 | Seaweed |
| Marseilleviridae | dsDNA | − | Isometric | 1 circular | 368 | Amoeba |
| Medioniviridae | ssRNA | ? | ? | ? | ? | Tunicates |
| Megabirnaviridae | dsRNA | − | Isometric | Linear, segmented | 7 | Fungi |
| Mesoniviridae | ssRNA | + | Spherical | 1 linear | 20 | Vertebrates |
| Metaviridae | ssRNA | − | RT‐spherical | 1 + segment | 4–10 | Fungi, plants, invertebrates |
| Microviridae | ssDNA | − | Isometric | 1 + circular | 4–6 | Bacteria, spiroplasmas |
| Mimiviridae | dsDNA | − | Isometric | 1 linear | 1200 | Amoeba |
| Mymonaviridae | ssRNA | + | Filamentous | 1 linear | 10 | Fungi |
| Myoviridae | dsDNA | − | Tailed phage | 1 linear | 39–169 | Bacteria, archaea |
| Nairoviridae | ssRNA | + | Spherical | 3 linear negative sense | 11–20 | Vertebrates, arthropods |
| Nanoviridae | ssDNA | − | Isometric | 6–9 circular | 6–9 | Plants |
| Narnaviridae | ssRNA | − | RNP complex | 1 + segment | 2–3 | Fungi |
| Nimaviridae | dsDNA | + | Ovoid | 1 circular | 293 | Crustaceans |
| Nodaviridae | ssRNA | − | Isometric | 2 + segments | 4–5 | Vertebrates and invertebrates |
| Nudiviridae | dsDNA | + | Rod‐shaped | 1 circular | 96–231 | Invertebrates |
| Nyamiviridae | ssRNA | + | Spherical | 1 linear | 11.6 | Birds, invertebrates |
| Orthomyxoviridae | NssRNA | + | Pleomorphic | 6–8 − segments | 10–15 | Vertebrates |
| Papillomaviridae | dsDNA | − | Isometric | 1 circular | 7–8 | Vertebrates |
| Paramyxoviridae | NssRNA | + | Pleomorphic | 1 − segment | 15 | Vertebrates |
| Partitiviridae | dsRNA | − | Isometric | 2 segments | 4–6 | Plants, fungi |
| Parvoviridae | ssDNA | − | Isometric | 1 +/− circular | 4–6 | Vertebrates and invertebrates |
| Peribunyaviridae | ssRNA | + | Spherical | 3, linear, negative sense | 11–20 | Humans, rodents, arthropods |
| Permutotetraviridae | ssRNA | − | Isometric | 1 linear | 5.6 | Invertebrates |
| Phasmaviridae | ssRNA | + | Spherical | 3, linear, negative sense | 10.3 | Insects |
| Phenuiviridae | ssRNA | + | Spherical | 3, linear, negative sense | 11–20 | Ruminants, camels, humans, mosquitos |
| Phycodnaviridae | dsDNA | − | Isometric | 1 linear | 160–380 | Algae |
| Picobirnaviridae | dsRNA | − | Isometric | 2 linear segments | 2.5 + 1.7 | Mammals |
| Picornaviridae | ssRNA | − | Isometric | 1 + segment | 7–8 | Vertebrates |
| Plasmaviridae | dsDNA | + | Pleomorphic | 1 circular | 12 | Mycoplasmas |
| Pleolipoviridae | ssDNA or dsDNA | + | Pleomorphic | 1 circular or linear | 7–16 | Archaea |
| Pneumoviridae | ssRNA | + | Spherical | 1 linear | 15 | Vertebrates |
| Podoviridae | dsDNA | − | Tailed phage | 1 linear | 19–44 | Bacteria |
| Polycipiviridae | ssRNA | − | Isometric | 1 linear + sense | 11 | Insects |
| Polydnaviridae | dsDNA | + | Rod, fusiform | Multiple supercoiled | 150–250 | Invertebrates |
| Polyomaviridae | dsDNA | − | Isometric | 1 circular | 5 | Vertebrates |
| Portogloboviridae | dsDNA | − | Isometric | 1 circular | 20 | Sulfolobus S38A archaea |
| Pospiviroidae | ssRNA | N/A | N/A | 1 circular | 0.24–0.4 | Plants |
| Potyviridae | ssRNA | − | Filamentous | 1/2 + segments | 8–12 | Plants |
| Poxviridae | dsDNA | + | Pleomorphic | 1 linear | 130–375 | Vertebrates and invertebrates |
| Pseudoviridae | ssRNA | − | RT‐spherical | 1 + segment | 5–8 | Fungi, plants, invertebrates |
| Qinviridae | ssRNA | ? | ? | ? | ? | ? |
| Quadriviridae | dsRNA | + | Spherical | Linear, 4 segments | 16.8 | Fungi |
| Reoviridae | dsRNA | − | Isometric | 10–12 segments | 19–32 | Vertebrates and invertebrates, plants |
| Retroviridae | ssRNA | − | RT + spherical | 1 dimer + segment | 7–12 | Vertebrates |
| Rhabdoviridae | NssRNA | + | Bullet‐shaped | 1 − segment | 11–15 | Vertebrates, plants |
| Roniviridae | ssRNA | + | Bacilliform | 1 linear | 26 | Crustaceans |
| Rudiviridae | dsDNA | + | Rod‐shaped | 1 linear | 33–36 | Archaea |
| Sarthroviridae | ssRNA | − | Isometric | 1 linear | 0.9 | Crustaceans |
| Secoviridae | ssRNA | − | Isometric | Linear/segmented | 24 | Plants |
| Siphoviridae | dsDNA | − | Tailed phage | 1 linear | 22–121 | Bacteria, archaea |
| Smacoviridae | ssDNA | − | Isometric | 1 circular | 2.3–2.8 | Vertebrates (?) |
| Sphaerolipoviridae | dsDNA | − | Isometric | 1 circular | 16–19 | Bacteria, archaea |
| Spiraviridae | ssDNA | − | Cylindrical | 1 circular | 25 | Archaea |
| Sunviridae | ssRNA | ? | ? | ? | 17 | Vertebrates |
| Tectiviridae | dsDNA | − | Isometric | 1 linear | 15 | Bacteria |
| Tobaniviridae | ssRNA | + | Spherical | 1 + segment | 28 | Vertebrates |
| Togaviridae | ssRNA | + | Isometric | 1 + segment | 10–12 | Vertebrates |
| Tolecusatellitidae | ssDNA | N/A | N/A | 1 | 0.7–1.35 | Plants |
| Tombusviridae | ssRNA | − | Isometric | 1/2 + segments | 4–5 | Plants |
| Totiviridae | dsRNA | − | Isometric | 1 segment | 5–7 | Fungi, protozoa |
| Tristromaviridae | dsDNA | + | Rod‐shaped | 1 linear | 15.9 | Archaea |
| Turriviridae | dsDNA | − | Isometric | 1 circular | 17.6 | Archaea |
| Tymoviridae | ssRNA | − | Isometric | 1 linear | 6.5–7 | Plants |
| Virgaviridae | ssRNA | − | Rod‐shaped | Linear, segmented, or nonsegmented | 3.3–6.5 | Plants |
| Wupedeviridae | ssRNA | ? | ? | ? | ? | Insects |
| Xinmoviridae | ssRNA | ? | ? | 1 linear, − sense | 12 | Mosquitoes |
| Yueviridae | ssRNA | ? | ? | − sense | ? | ? |
+ sense: Positive‐sense; − sense: negative‐sense; dsRNA: double‐stranded RNA; N/A: not applicable; NssRNA: negative‐sense single‐stranded RNA; RNP: ribonucleoprotein; RT: reverse transcriptase; ssRNA: single‐stranded RNA.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Basic Virology»
Представляем Вашему вниманию похожие книги на «Basic Virology» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Basic Virology» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












